Research
Biological interactions
Much of my scientific work is based on the concept of biological interactions, in which multiple factors (say, a genetic variant and some lifestyle behavior) synergistically affect a trait of interest (a risk factor for disease or the disease itself). This added complexity contributes to problems in biological and medical science, ranging from the “missing heritability” in genomics (why can’t our predictive models successfully account for the known heritability of many traits?) to the inconsistency of individual-specific responses in dietary trials. It also forms the basis for precision medicine by describing the ways that particular characteristics of an individual determine what behaviors, drugs, or environments will be most effective for them.
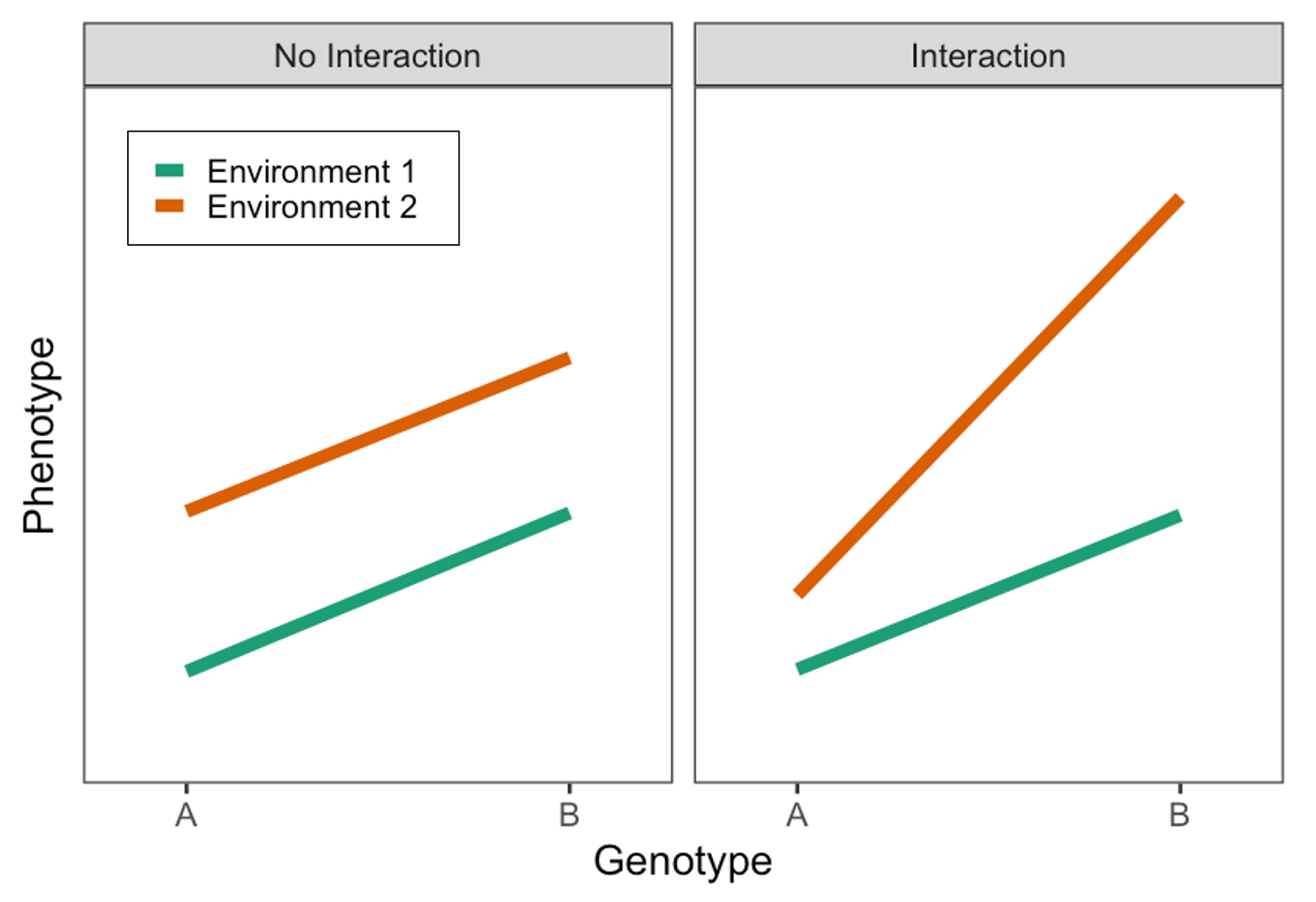
Most notably, I focus on gene-environment interactions (GxEs); here, the impact of a basic trait (e.g., biological sex), behavior (e.g., physical activity), or environmental exposure (e.g., pollution) on some outcome depends on one’s genetic makeup. We can also think about it as the exposure modifying the genetic effect on that outcome (this is equivalent from a statistical perspective!).
I’m grateful for the opportunity to collaborate on some of these problems with other experts in the GxE field, including Arun Durvasula, Alisa Manning, Han Chen, and others.
My GxE work falls into a few different domains:
Polygenic scores for interaction/response
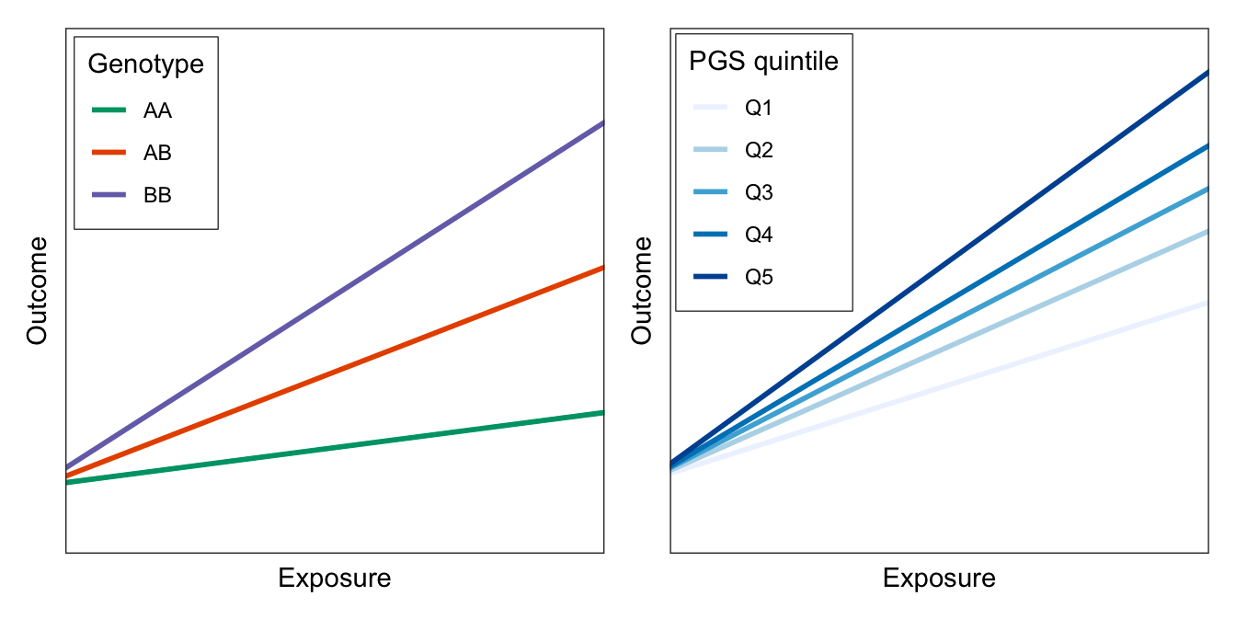
Single-variant GxEs are difficult to find and replicate because the effects are so small. Meanwhile, polygenic scores (PGS) are gaining popularity as a way to aggregate genetic information across the genome, but they are designed to predict risk of disease, rather than response to some therapeutic intervention. I have worked on multiple different methods for combining the concepts of PGS and GxE interactions to produce genetic scores that might help predict how people will respond differently to lifestyle changes of clinical treatments. One of these ideas involves aggregating over genome-wide GxE effects, rather than genome-wide main effects. Another involves testing for interaction at the level of biological pathways; we think that pathway-specific PGS might be more likely to show strong interactions than genome-wide (standard) PGS.
Molecular basis for diet response
Precision nutrition approaches to metabolic disease prevention will require a better understanding of the molecular factors governing inter-individual variability in diet response. To this end, my research has focused on examining how cardiometabolic risk factors are impacted by interactions between diet and molecular quantities, including genomics, epigenomics, and blood biomarkers. Using the Women’s Health Initiative Dietary Modification Trial dataset, I provided evidence that aggregation of thousands of genetic variants into a single “responder score” may predict the LDL-C response to dietary fat.
Two recent investigations have built on this by identifying gene-diet interactions impacting glycemic traits in larger study samples including the UK Biobank and TOPMed Consortium. One substantial challenge for studies like this is defining a dietary exposure that is both meaningful and measurable using data from dietary questionnaires. We took two different approaches to this problem in these studies: defining data-driven, food group-based dietary patterns and assessing macronutrient consumption (such as an exchange of fat and carbohydrate), respectively. Expanding beyond solely genetics, I collaborated with a colleague to conduct a pilot study that found an epigenetic signature of responders to vitamin K supplementation.
Large-scale, gene-environment interaction methods and discovery
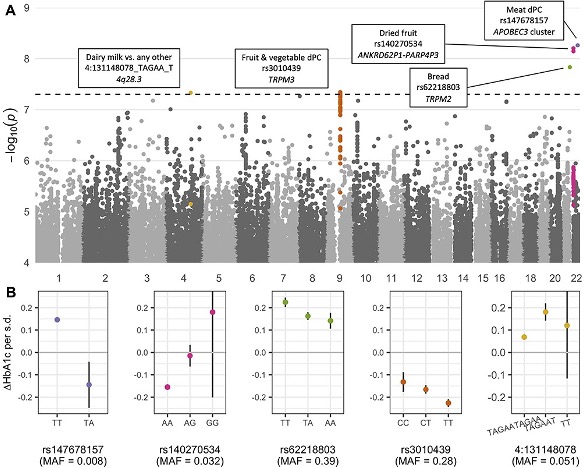
GxE identification provides a key foundation for genome-based precision medicine, but GxE analysis in increasingly larger samples carries statistical and computational challenges. I have made contributions towards overcoming these challenges in both methodology-focused and applied studies. I led the testing and publication of a tool, GEM, for more computationally efficient and statistically rigorous GxE testing, showing in the process that GEIs may be as common across the genome as genetic main effects.
Working closely with Joanne Cole, I put this program to work in conducting a novel “exposome-wide” GEI study in the UK Biobank that aims to systematically map interactions across many genetic variants, exposures, and metabolic biomarkers. Part of this effort involved searching across the genome for “variance-quantitative trait loci” (vQTLs), or genetic variants that associte with the variance (rather than the mean) of some trait, as a way to reduce the total amount of variants to be tested for GxE. The results from this project, including our catalog of vQTL and GxE summary statistics, are publicly available in a browser at the Common Metabolic Disease Knowledge Portal.
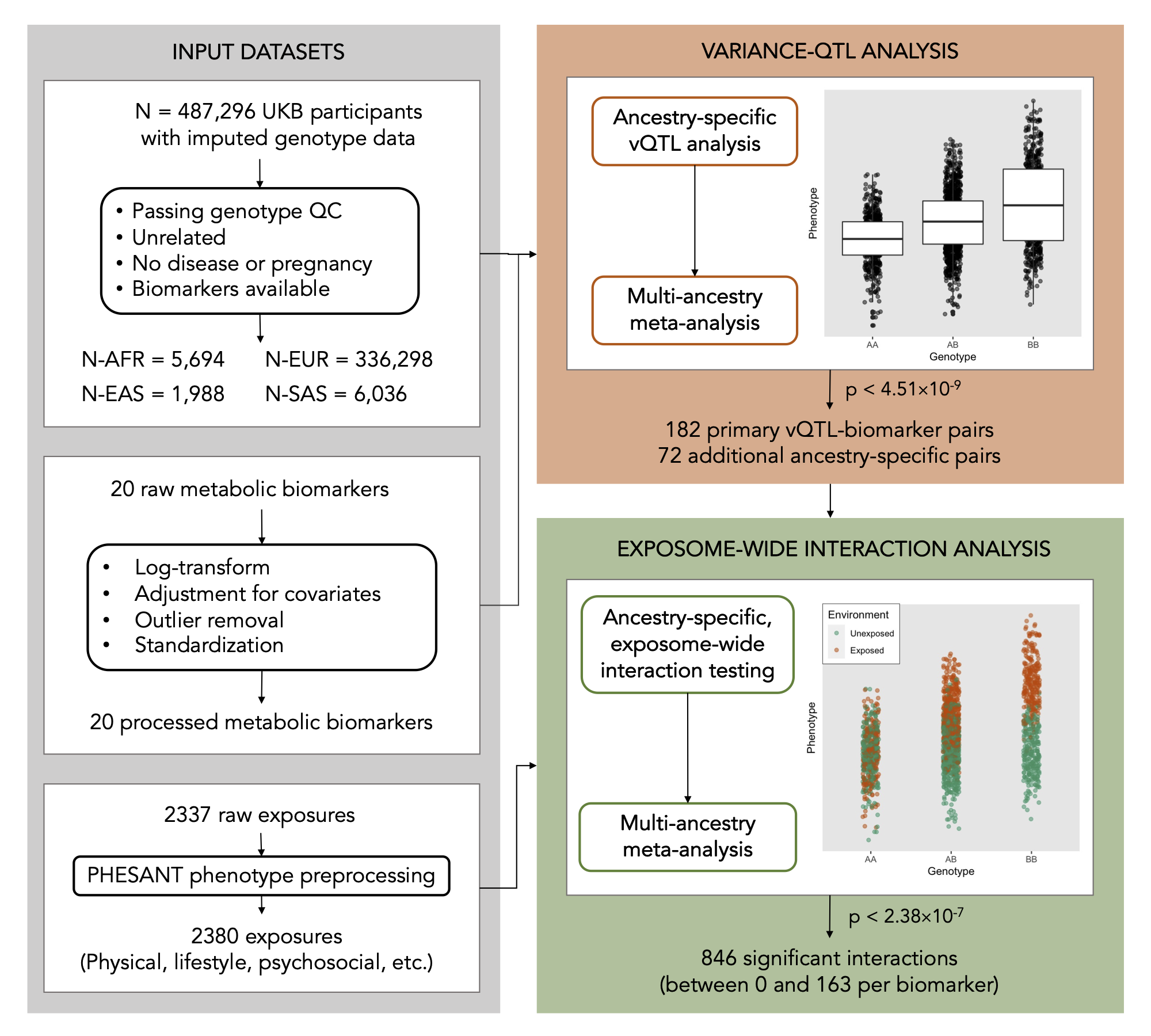
Molecular mediators of gene-environment interactions
GxEs will be better understood and more effectively translated into clinical relevance if we have a grasp of the underlying biological mechanisms. Molecular omics data, which are increasingly collected in biobanks and cohort studies, provide an opportunity to uncover some of these mechanisms.
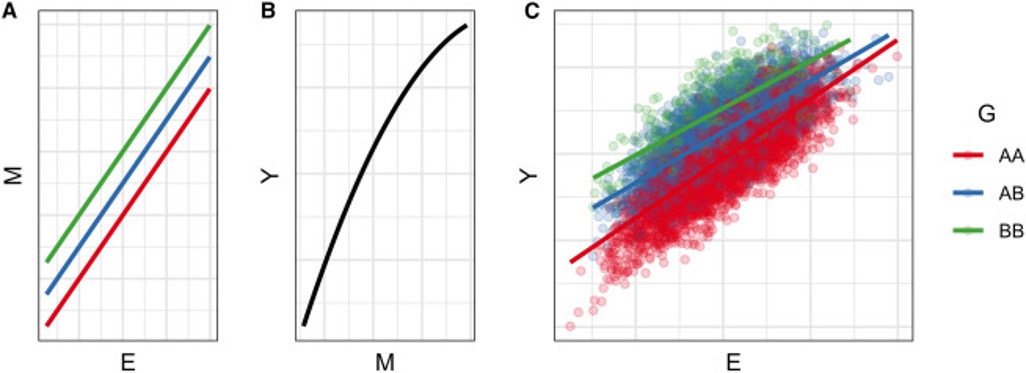
With Tamar Sofer, I published a review describing five mechanisms that occur at the molecular level that ultimately lead to a detectable GxE, many of which involve the action of a molecular mediator. An example mechanism is illustrated below, in which a GxE can be detected based on a combination of (1) linear genetic and environmental effects on a mediator and (2) a nonlinear relationship between that mediator and the outcome.
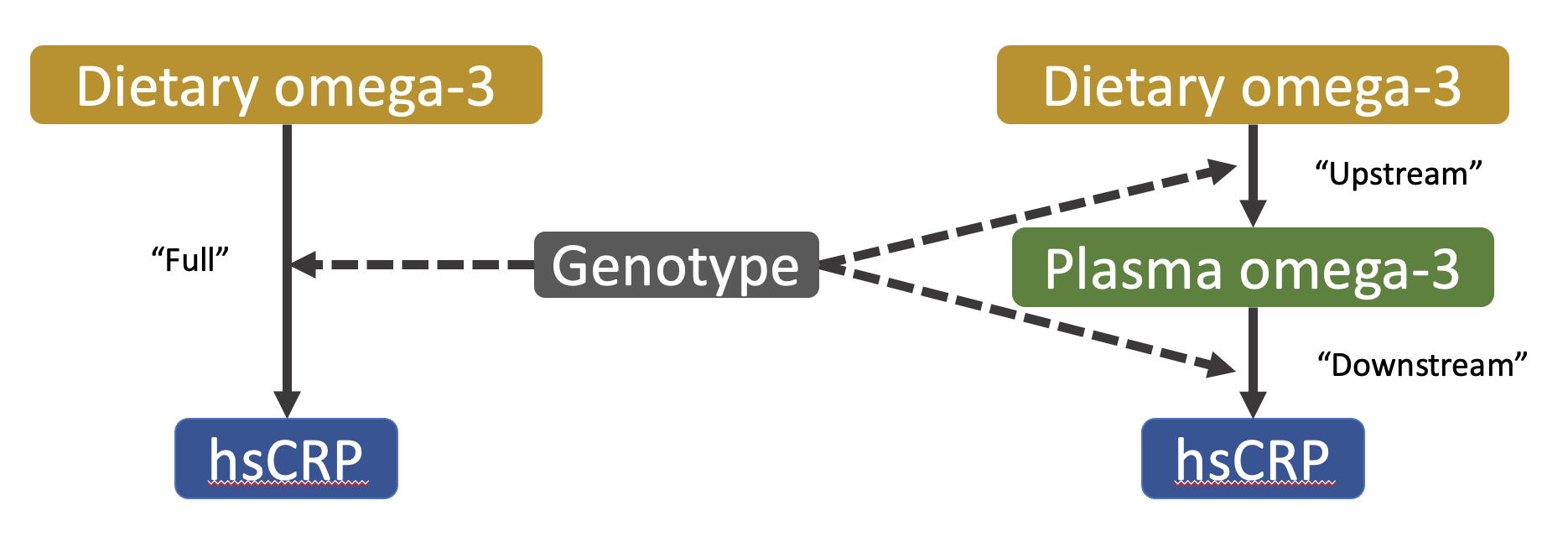
I have also explored the use of molecular data to improve the detection and understanding of GxEs through a “decomposition” approach, which sequentially tests for interaction upstream (genetic modification of the exposure-mediator relationship) and downstream (modification of the mediator-outcome relationship) of the mediator. A preliminary analysis in the UK Biobank has applied this approach, using plasma omega-3 fatty acids (N3FA) as a mediator of the relationship between dietary N3FA and chronic inflammation.
Additional topics
Epigenomics of cardiovascular disease risk
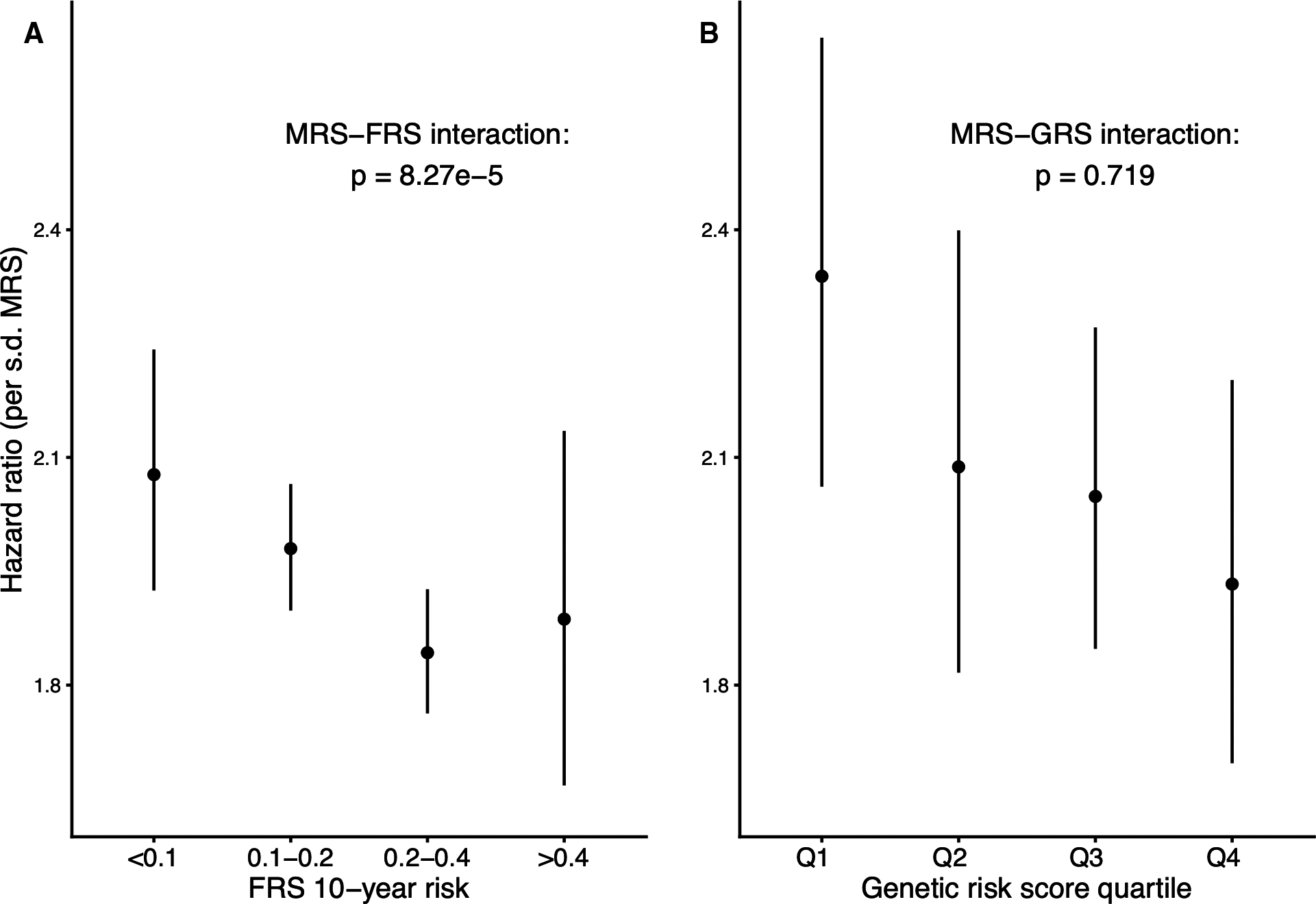
DNA methylation can integrate biological information from genetic variants and environmental inputs, and thus may act as a useful biomarker for cardiovascular disease. A series of investigations during my thesis work used bioinformatic approaches to explore connections between DNA methylation in blood cells and incident cardiovascular events in epidemiological cohorts. Beyond highlighting specific genes and pathways of interest, these studies provided important conceptual advances for the field of epigenetic epidemiology. Namely, we helped provide proof-of-concept for three ideas: (1) DNA methylation can act as a molecular readout of cumulative exposure to cardiovascular risk factors, (2) a methylation-based cardiovascular risk score may be differentially useful across strata of traditional or genetic risk, and (3) a recently introduced method for cross-study prediction of disease risk is viable in the novel context of cardiovascular epigenomics.
Genetic clustering
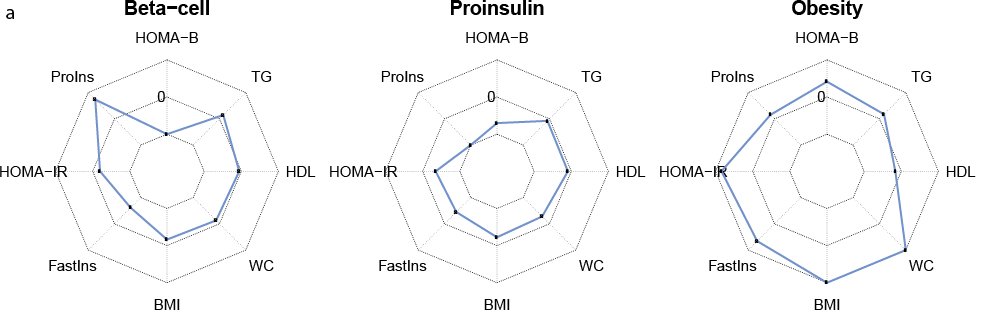
The sub-field of genetic clustering analysis aims to uncover disease subtypes based on genetic signatures of underlying disease-associated traits (“endophenotypes”). Miriam Udler has played a key role in pushing forward genetic clustering of type 2 diabetes using a Bayesian non-negative matrix factorization approach. I have helped generalize and partially automate the codebase for this clustering pipeline, which includes substantial preprocessing of genetic association summary statistics for these endophenotypes. This pipeline has been used in multiple follow-up papers on genetic clustering of type 2 diabetes, both introducing the pipeline and describing a multi-ancestry application.
Development of biomedical cloud computing platforms
Genomic science is increasingly migrating to cloud computing platforms for both scalability of computational resources and improved data security, but this migration process requires substantial infrastructure improvements. I have been fortunate to have opportunities to contribute to the development of these platforms. First, I have developed workflows (using Workflow Description Language) to conduct GxE tests with a variety of software tools in a way that is portable across different cloud platforms (e.g., Terra, DNAnexus, and NHLBI’s BioData Catalyst. Second, as a fellow in the BioData Catalyst Consortium, I led a monthly meeting with fellows and platform reps focused on workflow development (including their “accreditation”) and cloud analysis cost estimation.
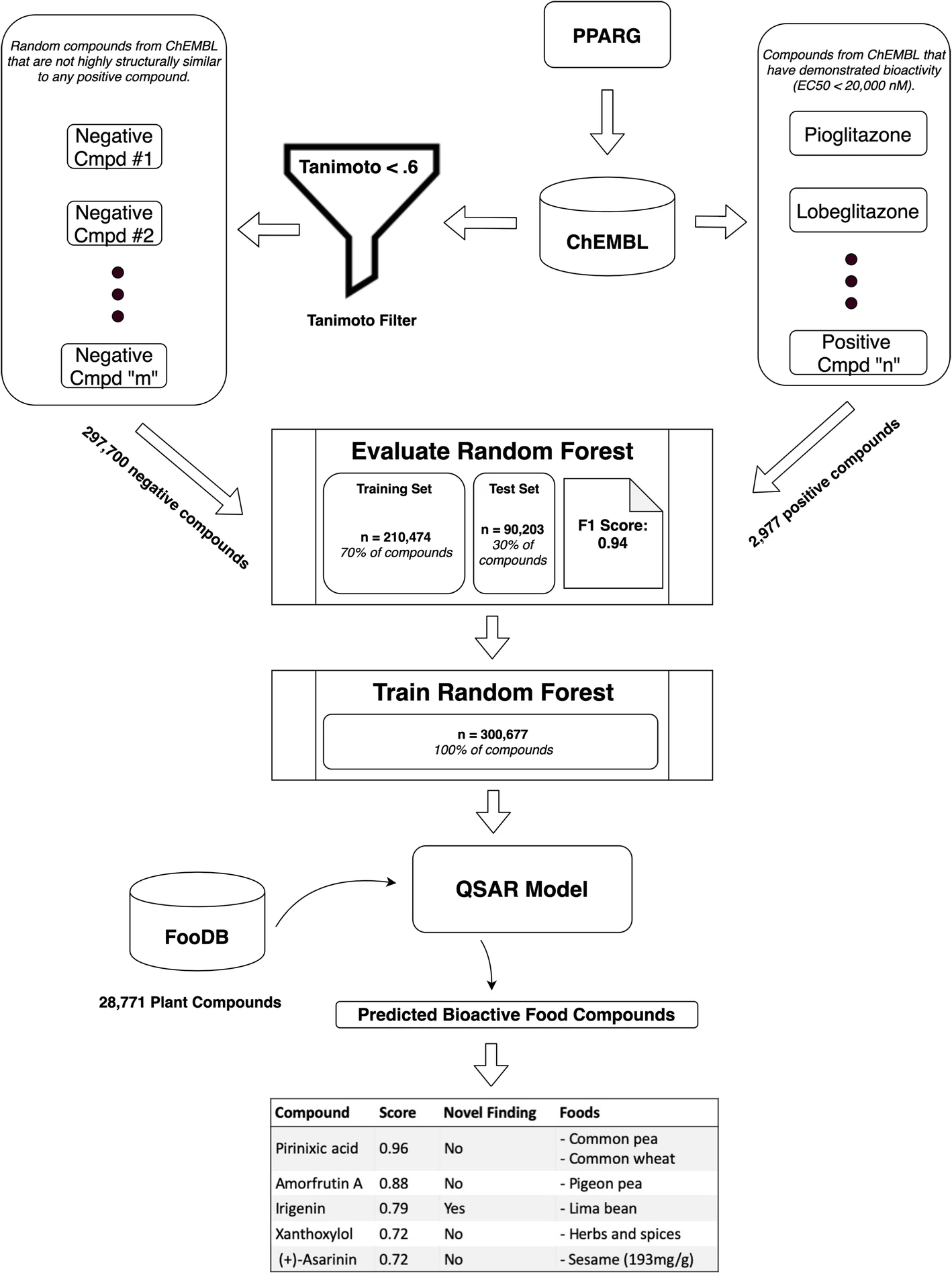
Prediction of dietary effects through food compound-drug similarity
Beginning in grad school, I explored a computational approach to predicting food compound bioactivity based on chemical similarity to drugs of known action. Since that point, I have worked with a small team to develop this idea and implement it as an open-source software tool called PhyteByte. This effort has not only prioritized food compounds of interest for specific phenotypes, but importantly helped translate established cheminformatics approaches from the pharmaceutical realm to the nutritional realm. We hope to expand this software by integrating better food compound databases and improved bioactivity prediction models (“Quantitative Structure-Activity Relationship” models). Great fun working with Sean Harrington and Larry Parnell on this project!